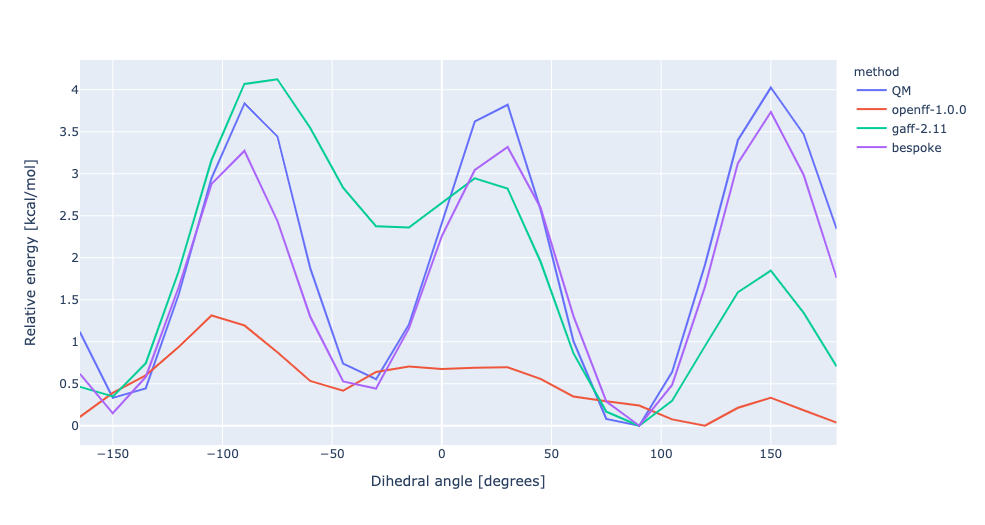
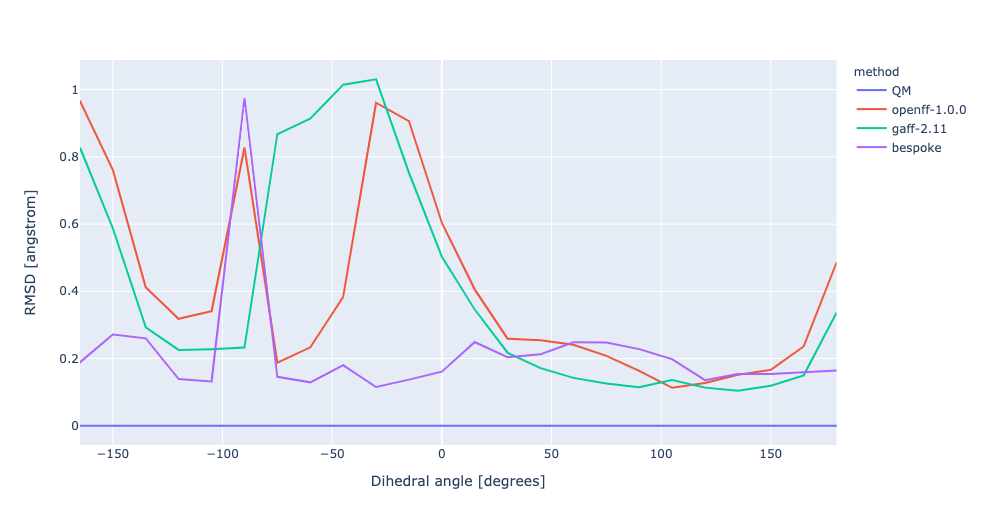
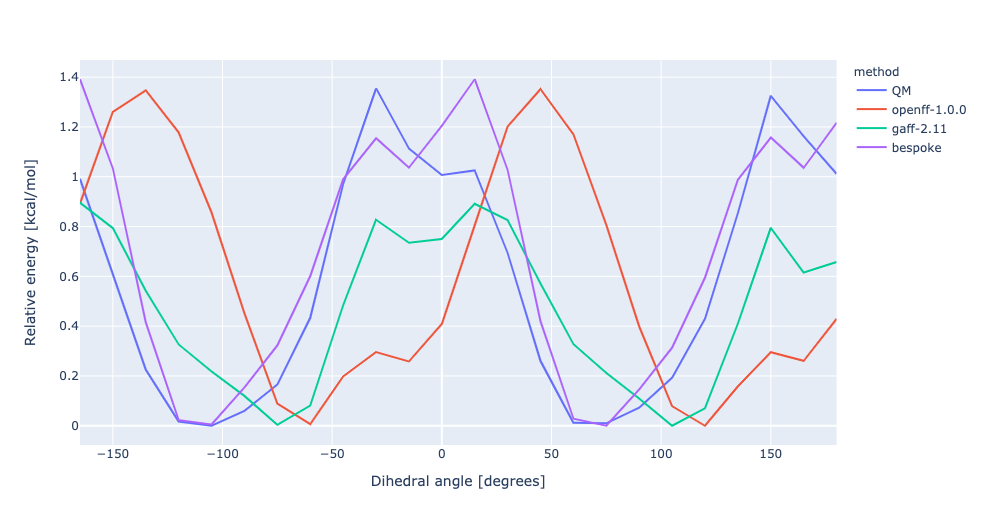
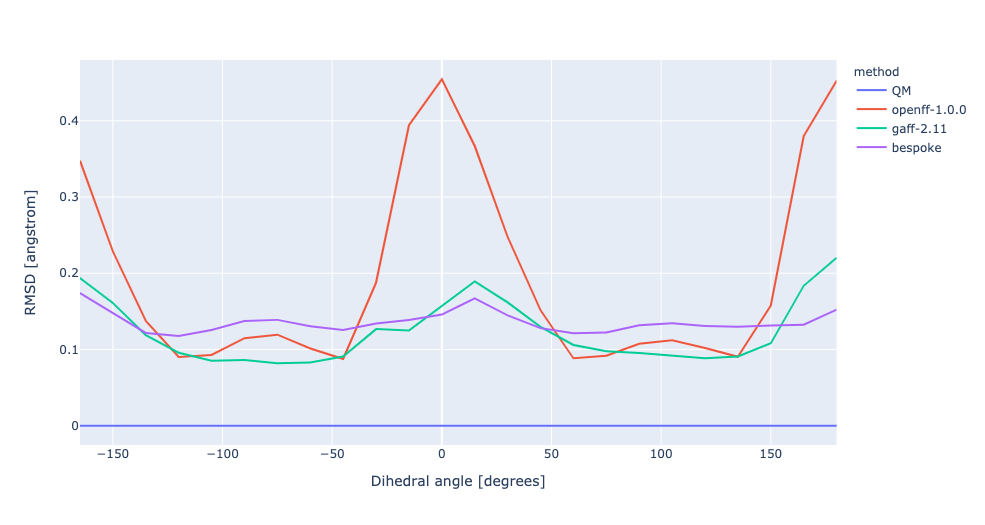
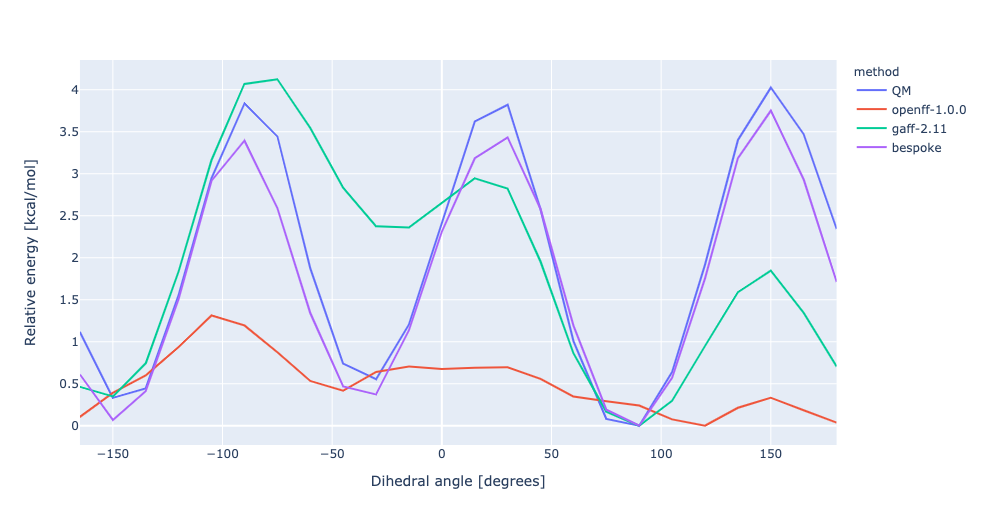
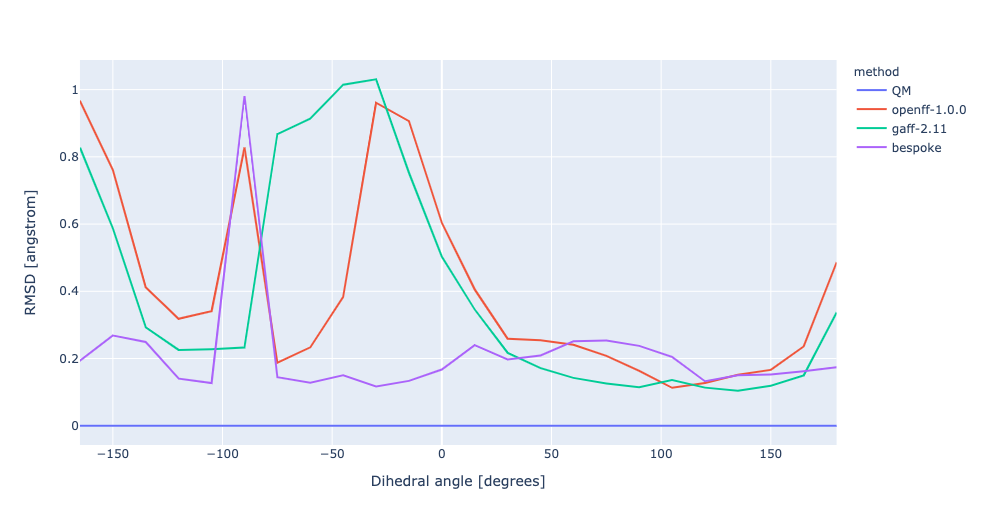
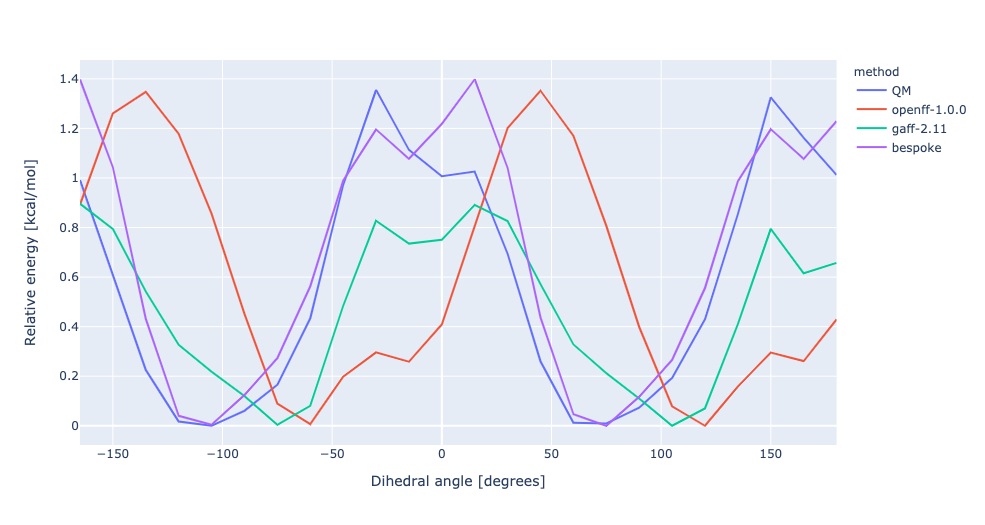
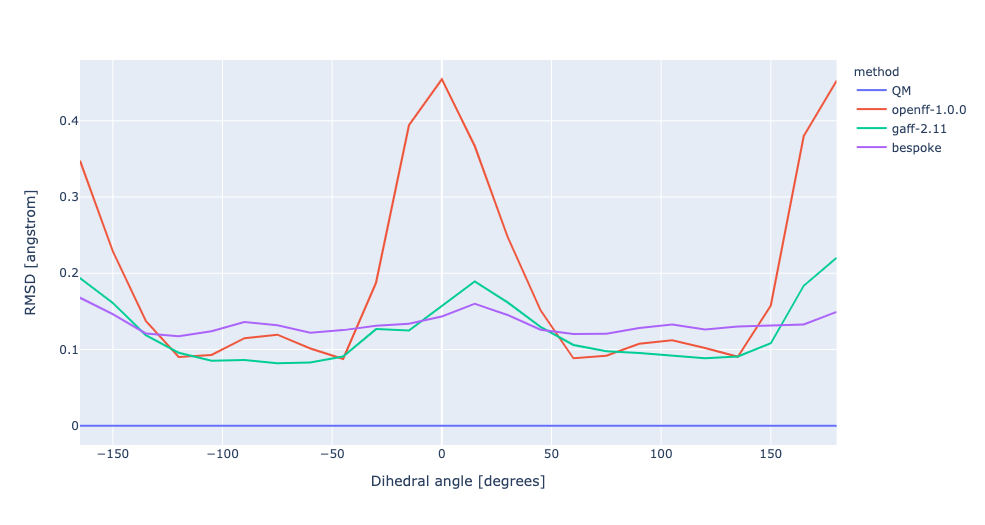
During torsion profile fitting we allow a slight MM relaxation of the molecule with 1kcal/mol restraints on all atoms not involved in the target dihedral. We recently looked into the effect of removing the restraints and for the small set of molecules saw a very similar performance in the resulting torsion profiles. However, we see much larger RMSD deviations during the fitting as the molecule fully relaxes. In this experiment, we have changed the torsion profile target to remove the restraints and use the RMSD deviations as part of the ForceBalance objective function. This should lead to both improved PES and lower RMSD between the MM and QM predicted structures.
Implementation
To do this we have looked at two possible similar methods that involve using the opt_geo target in combination with torsion profiles. This also gives us the ability to chose which internal degrees of freedom contribute to the objective function. In this example we only include dihedrals with a denom of 50, all other terms have a denom of 0.
Option 1, Set up a standard opt geo target for each grid point on the torsion drive with no restraints or frozen atoms. Fit these with each torsion profile.
Option2, Change the torsion profile target to compute the opt geo objective after each frozen optimization. This gives us a vector with the same length as the torsion profile where each value is the opt_geo dihedral only objective value, this is then added to the energy differences to give the total combined objective value.
return ((np.sqrt(self.wts)/self.energy_denom) * np.abs(compute.emm - self.eqm)) + ((np.sqrt(self.wts)/2 * total_ic_diff))
Option 1
This option was run by Trevor and the forcebalance tables will be updated later here we only have the validation torsion drives.
Restraints | Forcebalance result | Torsiondrive PES | Torsiondrive RMSD | RMSE |
|---|---|---|---|---|
No restraints | openff-1.0.0 RMSE 1.9340226158607 kcal/mol gaff-2.11 RMSE 1.1407243538497651 kcal/mol bespoke RMSE 0.3873825776904058 kcal/mol | |||
RMSD | openff-1.0.0 RMSE 1.9340226158607 kcal/mol gaff-2.11 RMSE 1.1407243538497651 kcal/mol bespoke RMSE 1.0285332638632987 kcal/mol |
Restraints | Forcebalance result | Torsiondrive PES | Torsiondrive RMSD | RMSE |
|---|---|---|---|---|
No restraints | openff-1.0.0 RMSE 0.7473961544204318 kcal/mol gaff-2.11 RMSE 0.3235153765035402 kcal/mol bespoke RMSE 0.1986501689347006 kcal/mol | |||
RMSD | openff-1.0.0 RMSE 0.7473961544204318 kcal/mol gaff-2.11 RMSE 0.3235153765035402 kcal/mol bespoke RMSE 0.3501012088022912 kcal/mol |
Option 2
State | Min(QM) | Min(MM) | Range(QM) | Range(MM) | Max-RMSD angstrom | Energy-RMSE kcal/mol |
|---|---|---|---|---|---|---|
initial (without restraints) | 90 | -45 | 0 - 4.027 | -0.123 - 2.616 | 0.606 | 0.9598 |
final (without restraints) | 90 | 90 | 0 - 4.027 | 0 - 3.907 | 0.277 | 0.2493 |
initial (with RMSD ) | 90 | -45 | 0 - 4.027 | -0.123 - 2.616 | 0.606 | 0.9598 |
final (with RMSD) | 90 | 90 | 0 - 4.027 | 0 - 3.915 | 0.274 | 0.1919 |
Restraints | Forcebalance result | Torsiondrive PES | Torsiondrive RMSD | RMSE |
|---|---|---|---|---|
No restraints | openff-1.0.0 RMSE 1.9340226158607 kcal/mol gaff-2.11 RMSE 1.1407243538497651 kcal/mol bespoke RMSE 0.3873825776904058 kcal/mol | |||
RMSD | openff-1.0.0 RMSE 1.9340226158607 kcal/mol gaff-2.11 RMSE 1.1407243538497651 kcal/mol bespoke RMSE 0.3468083675943567 kcal/mol |
State | Min(QM) | Min(MM) | Range(QM) | Range(MM) | Max-RMSD angstrom | Energy-RMSE kcal/mol |
|---|---|---|---|---|---|---|
initial (without restraints) | -105 | 135 | 0 - 1.355 | -1.058 - 0.640 | 0.206 | 1.2118 |
final (without restraints) | -105 | 60 | 0 - 1.355 | -0.132 - 1.192 | 0.170 | 0.1878 |
initial (with RMSD ) | -105 | 135 | 0 - 1.355 | -1.058 - 0.640 | 0.206 | 1.2118 |
final (with RMSD ) | -105 | 60 | 0 - 1.355 | -0.114 - 1.191 | 0.164 | 0.1711 |
Restraints | Forcebalance result | Torsiondrive PES | Torsiondrive RMSD | RMSE |
|---|---|---|---|---|
No restraints | openff-1.0.0 RMSE 0.7473961544204318 kcal/mol gaff-2.11 RMSE 0.3235153765035402 kcal/mol bespoke RMSE 0.1986501689347006 kcal/mol | |||
RMSD | openff-1.0.0 RMSE 0.7473961544204318 kcal/mol gaff-2.11 RMSE 0.3235153765035402 kcal/mol bespoke RMSE 0.19424683559308076 kcal/mol |
No regularization
Here we run the RMSD fit again and the no restraint standard fitting without regularization to compare the difference.
State | Min(QM) | Min(MM) | Range(QM) | Range(MM) | Max-RMSD angstrom | Energy-RMSE kcal/mol |
|---|---|---|---|---|---|---|
initial (without restraints) | 90 | -45 | 0 - 4.027 | -0.123-2.616 | 0.606 | 0.9598 |
final (without restraints) | 90 | 90 | 0 - 4.027 | 0-4.014 | 0.328 | 0.0117 |
initial (with RMSD ) | 90 | -45 | 0 - 4.027 | -0.123-2.616 | 0.606 | 0.9598 |
final (with RMSD) | 90 | 90 | 0 - 4.027 | 0 - 4.551 | 0.288 | 0.1499 |
Restraints | Forcebalance result | Torsiondrive PES | Torsiondrive RMSD | RMSE |
|---|---|---|---|---|
No restraints | openff-1.0.0 RMSE 1.9340226158607 kcal/mol gaff-2.11 RMSE 1.1407243538497651 kcal/mol bespoke RMSE 0.8106549613587883 kcal/mol | |||
RMSD | openff-1.0.0 RMSE 1.9340226158607 kcal/mol gaff-2.11 RMSE 1.1407243538497651 kcal/mol bespoke RMSE 0.24389724680495628 kcal/mol |
State | Min(QM) | Min(MM) | Range(QM) | Range(MM) | Max-RMSD angstrom | Energy-RMSE kcal/mol |
|---|---|---|---|---|---|---|
initial (without restraints) | -105 | 135 | 0 - 1.355 | -1.058-0.640 | 0.206 | 1.2118 |
final (without restraints) | -105 | -105 | 0 - 1.355 | 0-1.347 | 0.242 | 0.0237 |
initial (with RMSD ) | -105 | 135 | 0 - 1.355 | -1.058-0.640 | 0.206 | 1.2118 |
final (with RMSD ) | -105 | 60 | 0 - 1.355 | -0.112-1.124 | 0.159 | 0.1571 |
Restraints | Forcebalance result | Torsiondrive PES | Torsiondrive RMSD | RMSE |
|---|---|---|---|---|
No restraints | openff-1.0.0 RMSE 0.7473961544204318 kcal/mol gaff-2.11 RMSE 0.3235153765035402 kcal/mol bespoke RMSE 0.1834140590462866 kcal/mol | |||
RMSD | openff-1.0.0 RMSE 0.7473961544204318 kcal/mol gaff-2.11 RMSE 0.3235153765035402 kcal/mol bespoke RMSE 0.174320417515463 kcal/mol |