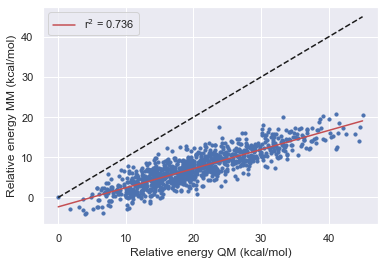
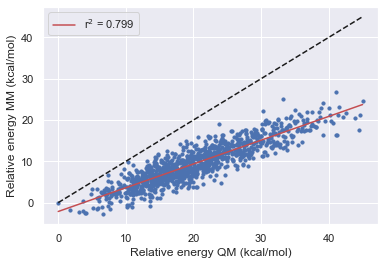
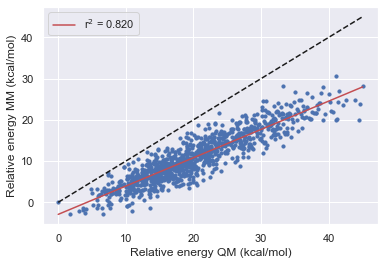
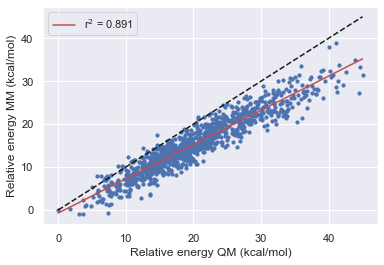
After using the bespoke workflow to optimize the torsions of some of the TYK2 ligands we want to establish how much the parameters have improved the overall description of the molecule. What's more, we also want to see how well the parameters fit molecule fragments transfer back to the parent molecule. In these tests, we hope to establish how the optimization has affected the force field before running FEP calculations.
Bespoke-fit
Each of the TYK2 molecules was fragmented using fragmenter. The WBO threshold was set to 0.03 and all non-rotor ring substituents were removed. A new smirks pattern was then made for each unique combination of 4 atoms running through the central bond fragmented around. 1D Torsion drives were then ran for each rotatable central bond on the public QCArchive using our default specification.
The smirks pattern included atoms up to 1 bond away from the 4 atoms in the dihedral to ensure that the patterns were transferable between fragments but were unique enough to not be miss applied. This is important as it ensures that smirks patterns are optimized in the presence of the parameters they will be used within production and allows self-consistent optimisation of interdependent torsions.
The Forcebalance target TorsionProfile was used with the L1 penalty. The upper energy limit was changed to 10 from 5 and the k value prior was set to 6. This is important as it allows the parameters to move sufficiently away from the starting values in cases where the original guess is poor. What's more due to the introduction of many more torsion terms (parsley uses just 4 k values compared to the 68 made in the bespoke workflow) this also stops the regularization term dominating the error and allows for a much better fit.
Molecule 7
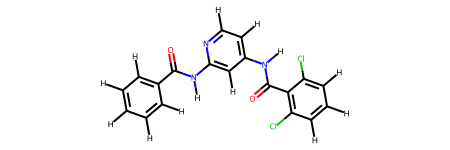
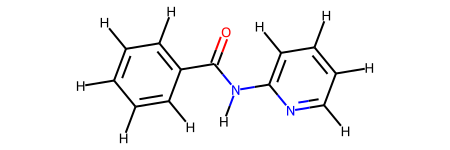
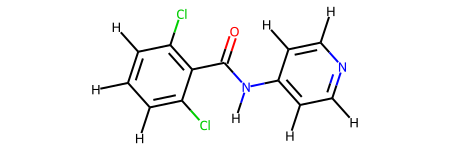
This molecule is a particularly challenging case for the bespoke torsion workflow due to the use of fragmentation resulting in two very similar molecules.



The difficulty here is that the bespoke smirks made for the fragments can easily be misapplied between the torsions. In this case, the bespoke torsion for the O=C-N-H torsion even with an extra layer of atoms is the same for both fragments. For example, the pattern is [#8AH0X1x0!r+0:1]=!@[#6AH0X3x0!r+0:2](-!@[#6aH0X3x2r6+0])-!@[#7AH1X3x0!r+0:3](-!@[#6aH0X3x2r6+0])-!@[#1AH0X1x0!r+0:4] and the the atoms matched by this in the fragments are shown below.

However, due to each unique dihedral passing through a central bond getting its own smirks the other dihedral terms are unique to the fragments and offer the extra degrees of freedom required to get a good bespoke fit for each fragment. In this case one of the other smirks is [#6aH0X3x2r6+0:1](:@[#6aH1X3x2r6+0])(:@[#6aH1X3x2r6+0])-!@[#6AH0X3x0!r+0:2](=!@[#8AH0X1x0!r+0])-!@[#7AH1X3x0!r+0:3](-!@[#1AH0X1x0!r+0])-!@[#6aH0X3x2r6+0:4](:@[#6aH1X3x2r6+0]):@[#7aH0X2x2r6+0] which hits the following atoms.

Validation
In order to validate any performance changes due to the refitting, a simple conformer energy ranking test was used. After running the bespoke workflow high-temperature MD was run on molecule 7 and each of the fitting fragments (using openff_unconstrained-1.0.0). 1000 confirmations were then extracted and the single point energies were calculated for all 3 molecules on a local QCArchive instance using multiple specifications (default(B3LYP-D3BJ/DZVP), openff-1.0.0/1.2.1/1.3.0 gaff-2.11 and ani2x, all mm specs were unconstrained.) The single point energies were then also recalculated using the bespoke forcefield output at the end of the bespoke workflow.
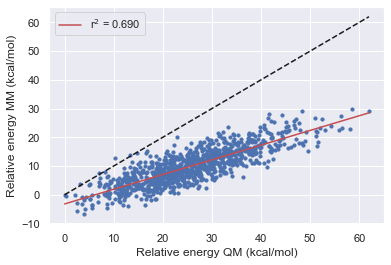
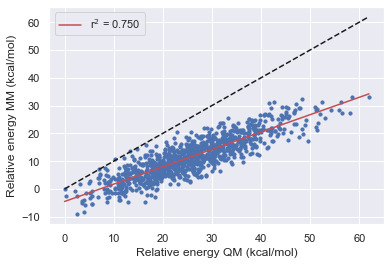
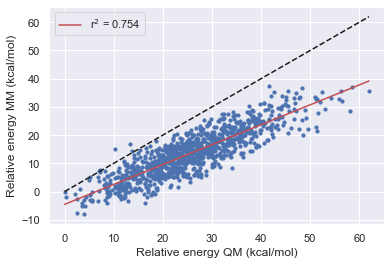
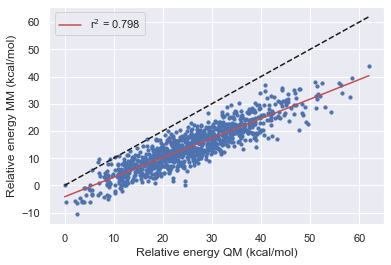
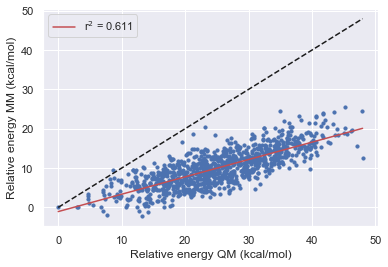
The Improvement of Openff
Here we see that OpenFF has consistently improved with each release in this test however Gaff-2.11 has the best performance across all of the molecules.
All force fields agree with QM for the parent molecule on the lowest energy structure.
All force fields disagree with QM on fragment 1 on the lowest energy structure
OpenFF-1.0.0 is the only force field to disagree with QM on the lowest energy structure this is however corrected in 1.2.1 and 1.30.
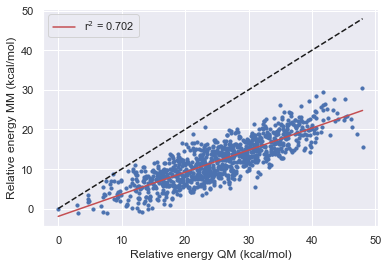
Bespoke-fit set 1
Here we look at the performance of the bespoke-fit forcefield starting from each of the Openff generations.
We see that the optimizations of torsions alone in the bespoke workflow are able to consistently improve the performance of the base-line force field.
Optimization of torsions alone seems to give around the same performance improvement as a forcefield release.
Improvements in the fragment performance seem to transfer well into the parent molecule.
Other factors such as bond/angle and improper parameters are also affecting the performance.
can we get further performance improvements by also refitting improper torsions directly linked to the targeted rotatable bond?
openff-1.3.0 seems to already be very good for these molecules with respect to the torsion terms as we see a negligible improvement with the torsion optimization.
we could also see some improvement by also fitting to the optimized geometries and vibrational frequencies of the fragments.
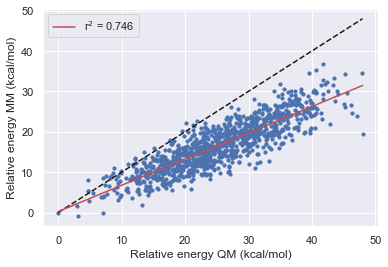
Validation set 2
Here we performed the same validation as above but this time starting from a trajectory again extracted from high temp MD but using the Gaff-2.11 forcefield. Due to the gaff barrier highlights being more accurate than that of Parsley we see that the range of relative energies is much smaller as we are trapped in a local minimum so this may need to be repeated from several starting locations.
Bespoke-fit set 2
Here we look at the performance of the bespoke-fit forcefield starting from each of the Openff generations and the second test set extracted from high-temperature MD using Gaff-2.11.
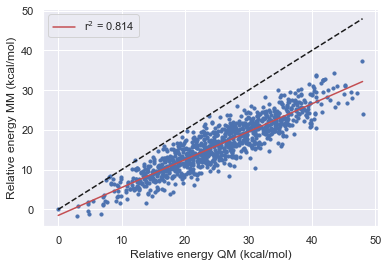
Validation set 3
Here we performed the same validation as above but this time starting from a trajectory extracted from an MD simulation with the k values for all rotatable torsions set to 0. This should allow the simulation to cross high energies barriers easily without any basis to the starting conformation.
Here we again see a large energy range for all of the molecules similar to set 1
We also see a general increase in accuracy with each version of the openff forcefields
All of the forcefields disagree with QM on the lowest energy structure in the validation sets
the performance of gaff-2.11 is comparable to openff-1.3.0 for the parent and fragment 1, but gaff is better for fragment 2
Using conformer specific charges set 2
I have also run the test on the gaff-2.11 generated conformers but used charges calculated specifically for the conformer.
The performance is generally worse as the forcefield is not designed to be used this way and was not fit with this in mind
Overall this seems to cause the forcefield to overestimated the relative energy of a conformer
This does help with fragment 2 where we see the forcefield disagree with QM on the lowest energy conformer.
Molecule | Openff-1.0.0 | Openff-1.2.1 | Openff-1.3.0 | Average Charges | Charge std |
|---|
 |  |  |  |  |  |
 |  |  |  |  |  |
 |  |  |  |  |  |
Using The TorsionDrive Trajectory
In the above fits, it was noted that future improvements in the performance could probably be hand by optimizing the bond and angle terms which were untouched during the first bespoke fit. One idea is to use the optimization trajectories extracted from the torsiondrive and fit to the relative energies of these conformers using the Abinitio forcebalance target. Following this, I have extracted the ~1700 single point calculations that make up the final torsion drive profile originally used for fitting and combined both fitting targets together. Here we allow the bond and angle force constants to optimize along with the bespoke torsion terms which are introduced as part of the bespoke workflow. Note that no bespoke bond and angle terms were introduced the parsley terms were just allowed to optimize.
We see that the performance of the model is increased but is not much better than just fitting the torsions in terms of 
We see that the MM relative energies are more consistently underestimated compared to QM after including the relative energy ranking.
Despite a very good fit in forcebalance this does not seem to transfer to these conformations from high-temperature MD.
Ani2x
Recently ani2x was used to correct free energy calculations and it was believed it was due to ani correcting the torsion potentials. To investigate this I have also performed the energy ranking test using ani2x.
Ani2x has the best correlation with our default spec, and would be a suitable replacement to optimize our MM torsion potentials to
Would we need to achieve a similar correlation in this test to see the same improvement in FEP as using Ani2x?